Pt Nanoparticles on MoS2(001) Surface via Adatoms.¶
Introduction.¶
This tutorial demonstrates how to create a platinum island on MoS2 by sequentially adding Pt adatoms, following the methodology described in the literature.
Manuscript
Saidi, W. A. "Density Functional Theory Study of Nucleation and Growth of Pt Nanoparticles on MoS2(001) Surface" Crystal Growth & Design, 15(2), 642–652. (2015) DOI: 10.1021/cg5013395. 12345
We will recreate the Pt island structure shown in Figure 4b:
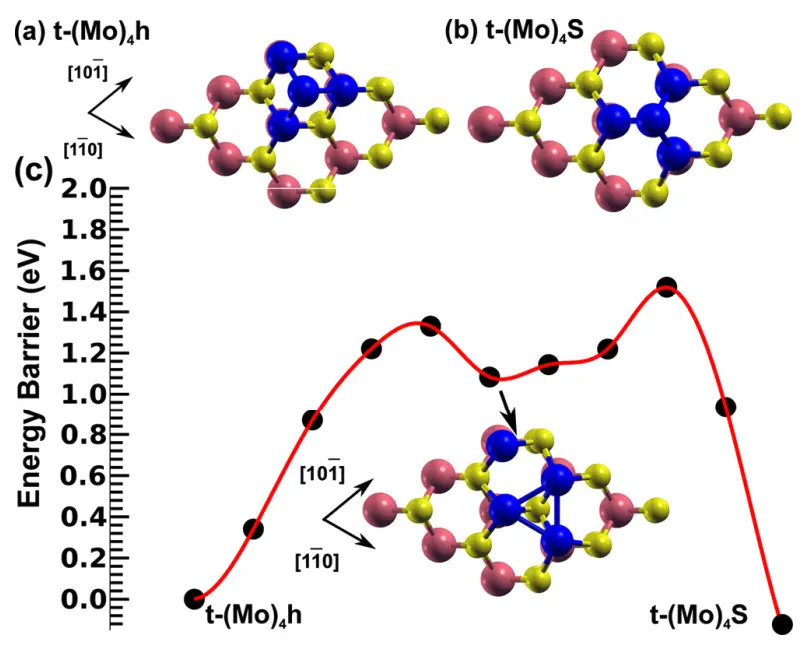
1. Create MoS2 Substrate.¶
1.1. Load Base Material.¶
Navigate to Materials Designer and import the MoS2 2D material from Standata.
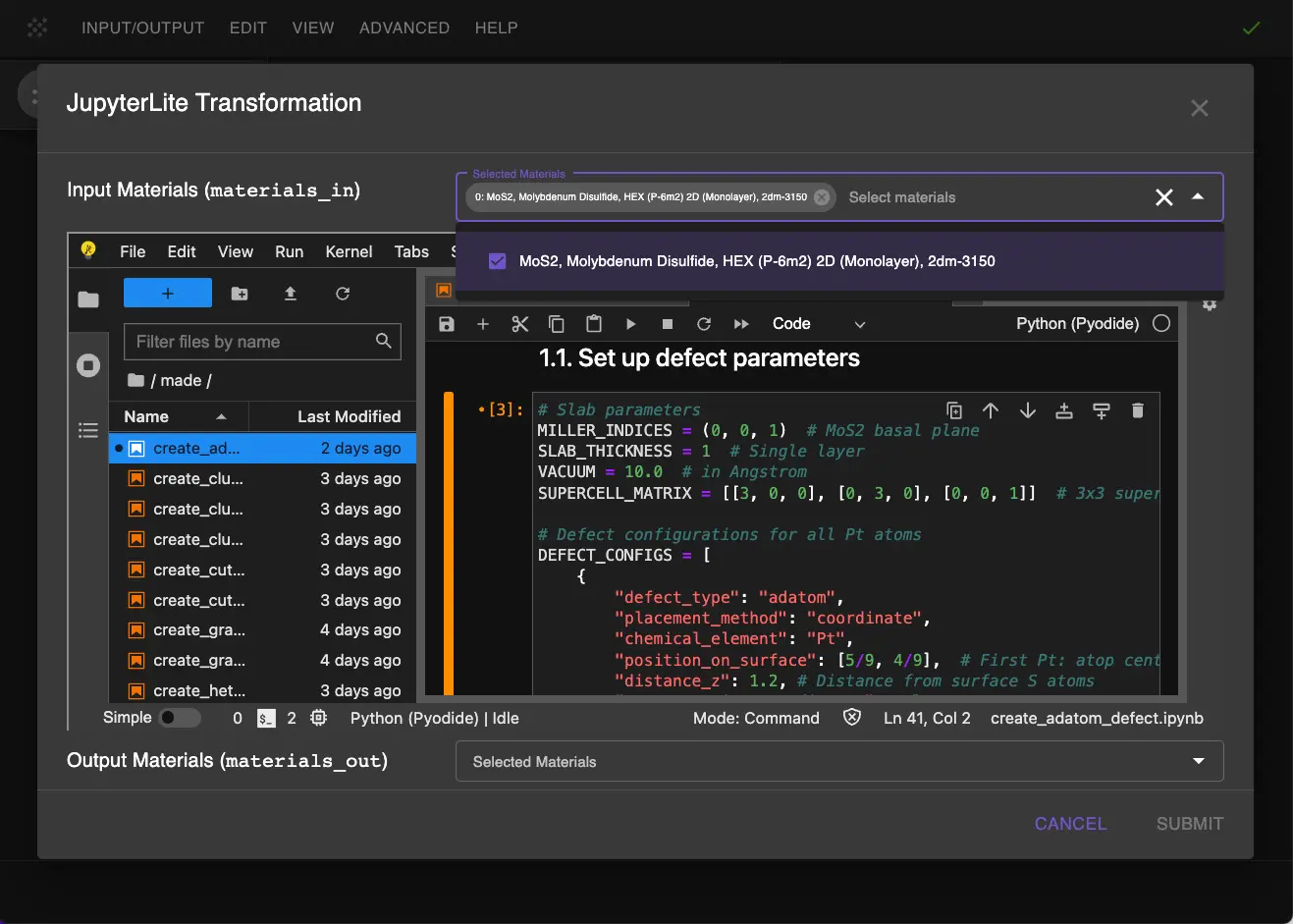
1.2. Launch JupyterLite Session.¶
Select the "Advanced > JupyterLite Transformation" menu item to launch the JupyterLite environment.
1.3. Open create_adatom_defect.ipynb Notebook.¶
Find and open the create_adatom_defect.ipynb notebook. Select MoS2 as input material.
2. Configure and Create Structure.¶
2.1. Set Parameters.¶
Set up the slab and defect parameters in the notebook:
# Index in the list of materials, to access as materials[MATERIAL_INDEX]
MATERIAL_INDEX = 0
ELEMENT = "Pt" # Chemical element of the adatom
# Dictionaries are validated and converted to AdatomDefectDict objects below
DEFECT_CONFIGS = [
{
"type": "adatom",
"coordinate_2d": [5/9, 4/9], # Crystal coordinates on the surface (x, y)
"distance_z": 1.2, # Method to place the adatom
"element": ELEMENT,
},
{
"type": "adatom",
"coordinate_2d": [2/9, 4/9], # Crystal coordinates on the surface (x, y)
"distance_z": 1.2, # Method to place the adatom
"element": ELEMENT,
},
{
"type": "adatom",
"coordinate_2d": [5/9, 7/9], # Crystal coordinates on the surface (x, y)
"distance_z": 1.2, # Method to place the adatom
"element": ELEMENT,
},
{
"type": "adatom",
"coordinate_2d": [4/9, 5/9], # Crystal coordinates on the surface (x, y)
"distance_z": 1.6, # Method to place the adatom
"element": ELEMENT,
},
]
PLACEMENT_METHOD = "new_crystal_site" # Method to place the adatom, e.g., "new_crystal_site", "exact_coordinate", "equidistant"
# Slab parameters
MILLER_INDICES = (0, 0, 1) # Miller indices of the surface
SLAB_THICKNESS = 1 # Thickness of the slab in unit cells
VACUUM = 10.0 # Vacuum thickness in Angstrom
XY_SUPERCELL_MATRIX = [[3, 0], [0, 3]] # Supercell matrix for the slab
TERMINATION_FORMULA = None # Stoichiometric formula of the slab termination to be used.
Key parameters explained:
-
Base layer Pt atoms (first three configs):
-
Positioned atop Mo atoms in a triangular arrangement
- Height 1.2 Å from surface S atoms to achieve separation from Mo atoms of 2.8 A -- from publication
-
Top Pt atom (fourth config):
-
Centered above the triangle, atop S atom
-
Height 1.6 Å from base Pt atoms (as they become the new surface layer)
-
distance_zsets the distance along the z-axis from the adatom to the topmost atom directly below it
2.2. Run the Notebook.¶
Execute the notebook to create the Pt island structure on MoS2 by selecting "Run" > "Run All Cells" from the JupyterLite menu.
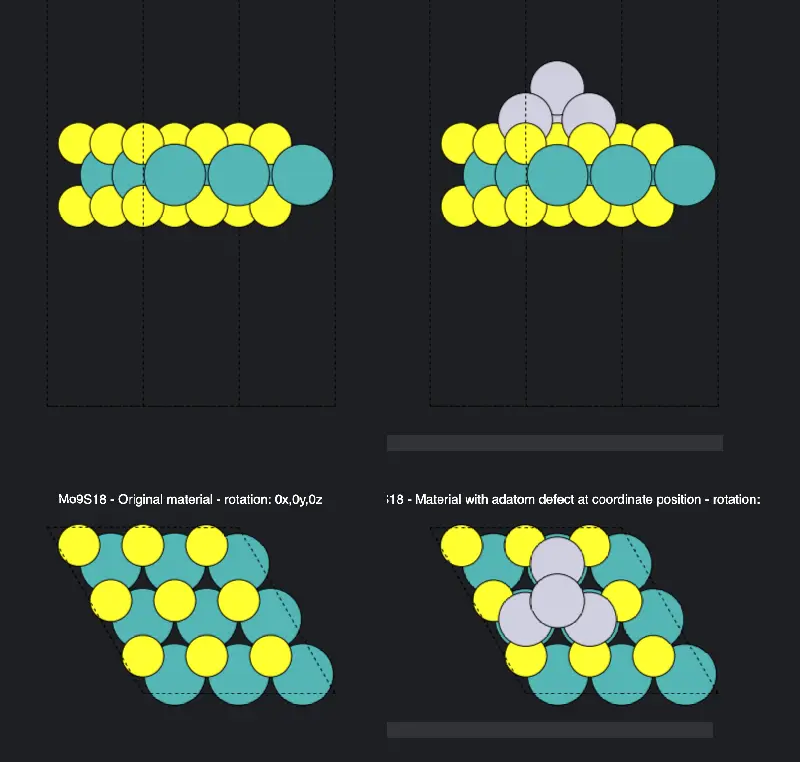
2.3. Pass the Result to Materials Designer.¶
The result can be passed to Materials Designer for visualization and viewed from the top:
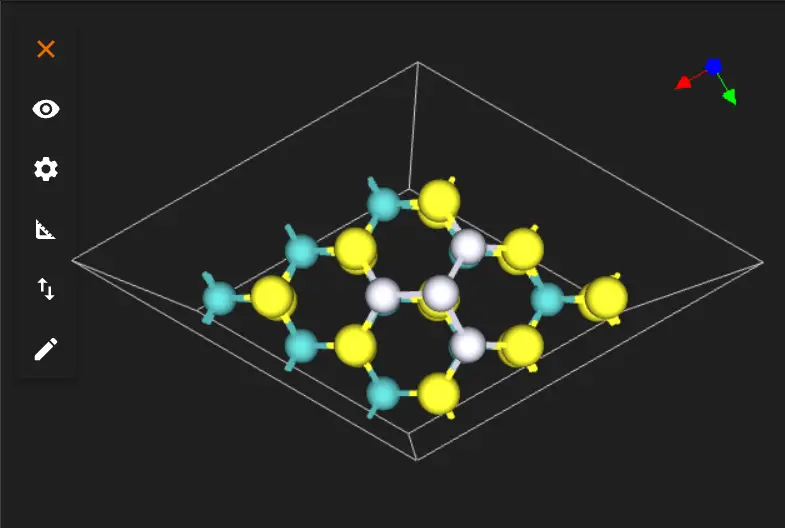
And from the side:
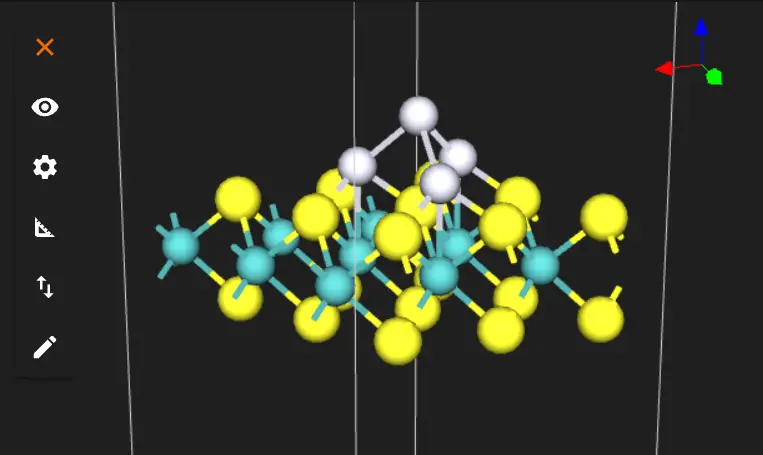
3. Analyze the Structure.¶
After adding all Pt atoms, verify the following:
3.1. Base Layer Geometry.¶
- Three Pt atoms should form a triangular base
- Each base Pt should be positioned atop Mo atoms
- Distance from surface S atoms should be ~1.2 Å
- Relaxation is needed to achieve the exact geometry from the publication, can be performed elsewhere
3.2. Top Atom Position.¶
- Fourth Pt should be centered above the triangle
- Position should be approximately above a surface S atom
- Height should be ~2.8 Å from surface (1.6 Å from base Pt atoms)
4. Save the Structure.¶
The final structure will be automatically passed back to Materials Designer where user can:
- Save it on the platform
- Export it in various formats
- Use it for further transformations
Interactive JupyterLite Notebook.¶
The following embedded notebook demonstrates the complete process. Select "Run" > "Run All Cells".
Parameter Fine-tuning.¶
To adjust the island structure:
- Base Layer:
- Adjust
APPROXIMATE_POSITION_ON_SURFACEto shift Pt positions -
Modify
DISTANCE_Zto change height from surface -
Top Atom:
- Adjust position to change island shape
- Modify height to change Pt-Pt spacing
References.¶
-
W. A. Saidi. Density functional theory study of nucleation and growth of pt nanoparticles on mos2(001) surface. Crystal Growth & Design, 15(2):642–652, 2015. URL: https://doi.org/10.1021/cg5013395. ↩
-
M. Jiao, W. Song, H.-J. Qian, Y. Wang, Z. Wu, S. Irle, and K. Morokuma. Qm/md studies on graphene growth from small islands on the ni(111) surface. Nanoscale, 8(5):3067–3074, 2016. URL: https://doi.org/10.1039/c5nr07680c. ↩
-
Kristen A. Fichthorn and Matthias Scheffler. Island nucleation in thin-film epitaxy: a first-principles investigation. Phys. Rev. Lett., 84:5371, 2000. URL: https://link.aps.org/doi/10.1103/PhysRevLett.84.5371. ↩
-
Jörg Neugebauer and Matthias Scheffler. Mechanisms of island formation of alkali-metal adsorbates on al(111). Phys. Rev. Lett., 71:577, 1993. URL: https://link.aps.org/doi/10.1103/PhysRevLett.71.577. ↩
-
Mahbube Hortamani, Peter Kratzer, and Matthias Scheffler. Density-functional study of mn monosilicide on the si(111) surface: film formation versus island nucleation. Phys. Rev. B, 76:235426, 2007. URL: https://link.aps.org/doi/10.1103/PhysRevB.76.235426. ↩